ProteinMPNN for Structure-based Design
ProteinMPNN is a GNN-based AI method for designing a protein sequence given a protein structure, a process known as structure-based design or inversefolding. ProteinMPNN uses various parameters to assist with protein design, including the temperature parameter, which controls diversity in the designs: values from 0.1 to 0.5 result in less diverse designs, while values of 0.5 and above lead to more diverse outcomes. LigandMPNN is coming soon.
How to Use ProteinMPNN
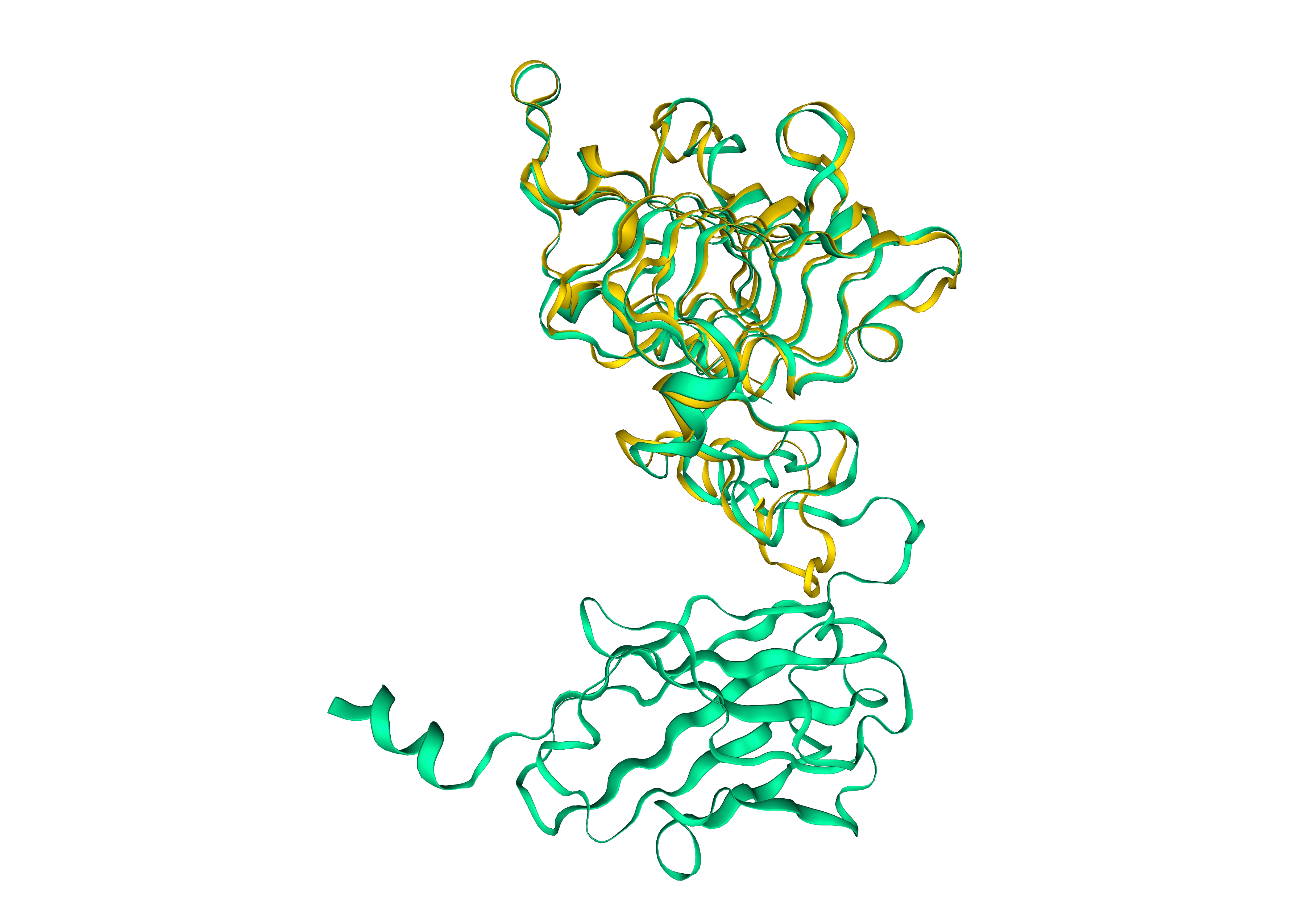
Use ProteinMPNN to design P00698
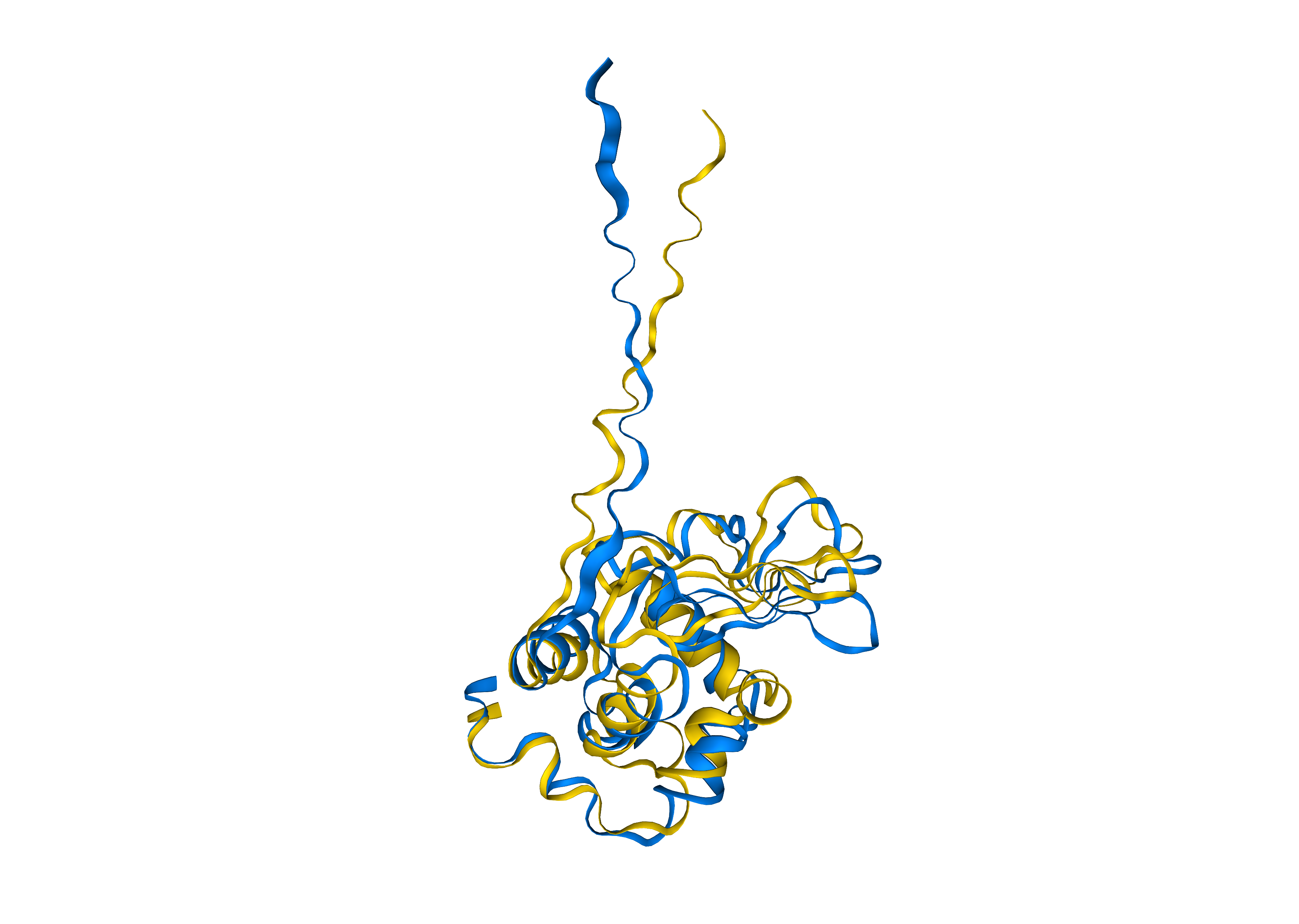
Inputs
- PDB file
A protein structure. Note this version only handles single chains. Updates coming soon.
Outputs
- Protein sequence
- A designed sequence of the same length as the input structure.
Examples
Use ProteinMPNN to design P00698 with a temperature of 1.0 and show 3 results
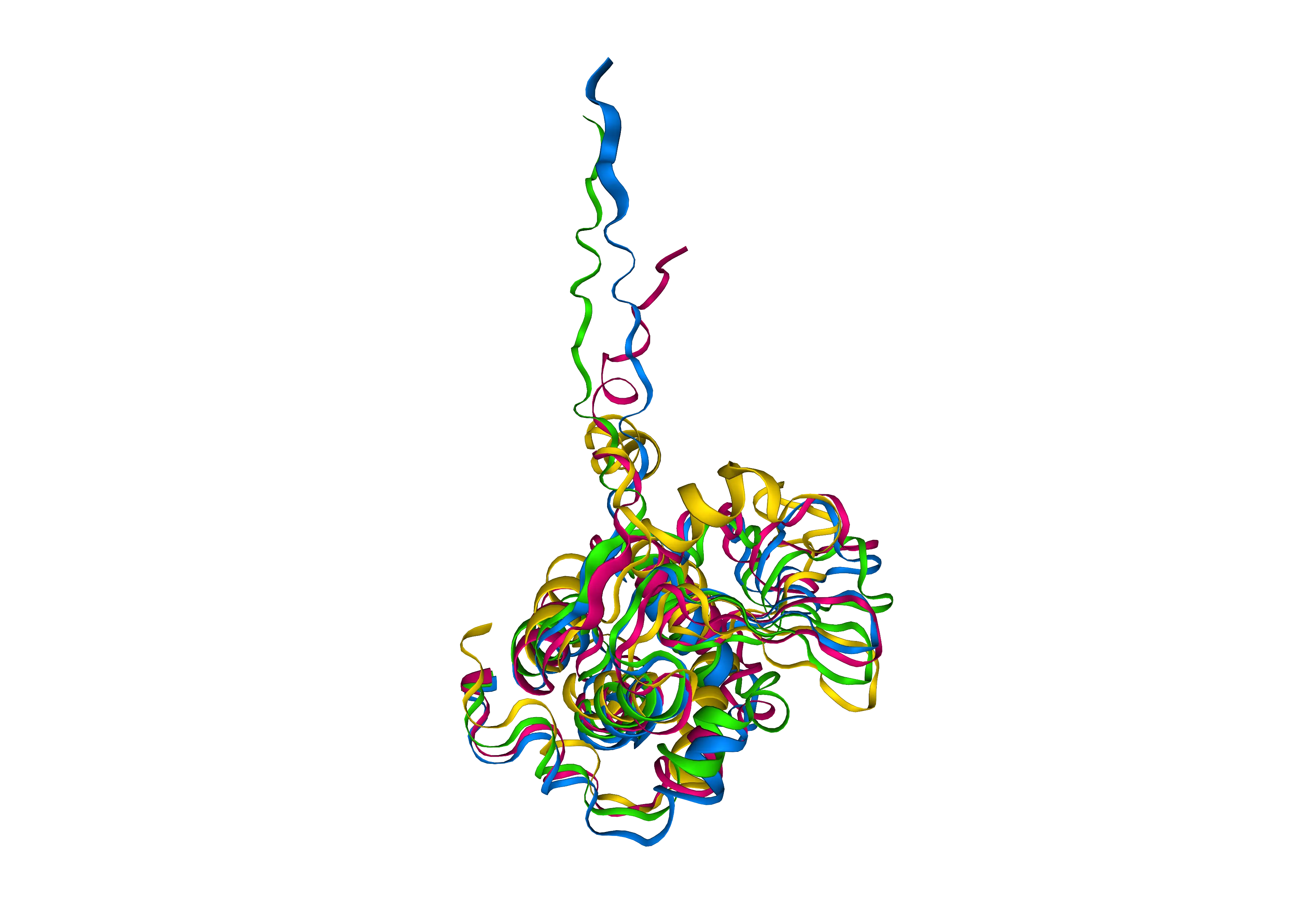
Use ProteinMPNN to design chain A of 4KRL with a temperature of 1.0, and fix all residues of chain B of 4KRL that are part of the interface with chain A