ESMFold for Protein Folding
ESMFold is an ML model that is a variation of AlphaFold2 that offers higher speed.
ESMFold Examples and Folding Scripts
Fold ‘DIHICGICKQQFNNLDAFVAHKQSGSQ’

Fold A0A7S7MT40
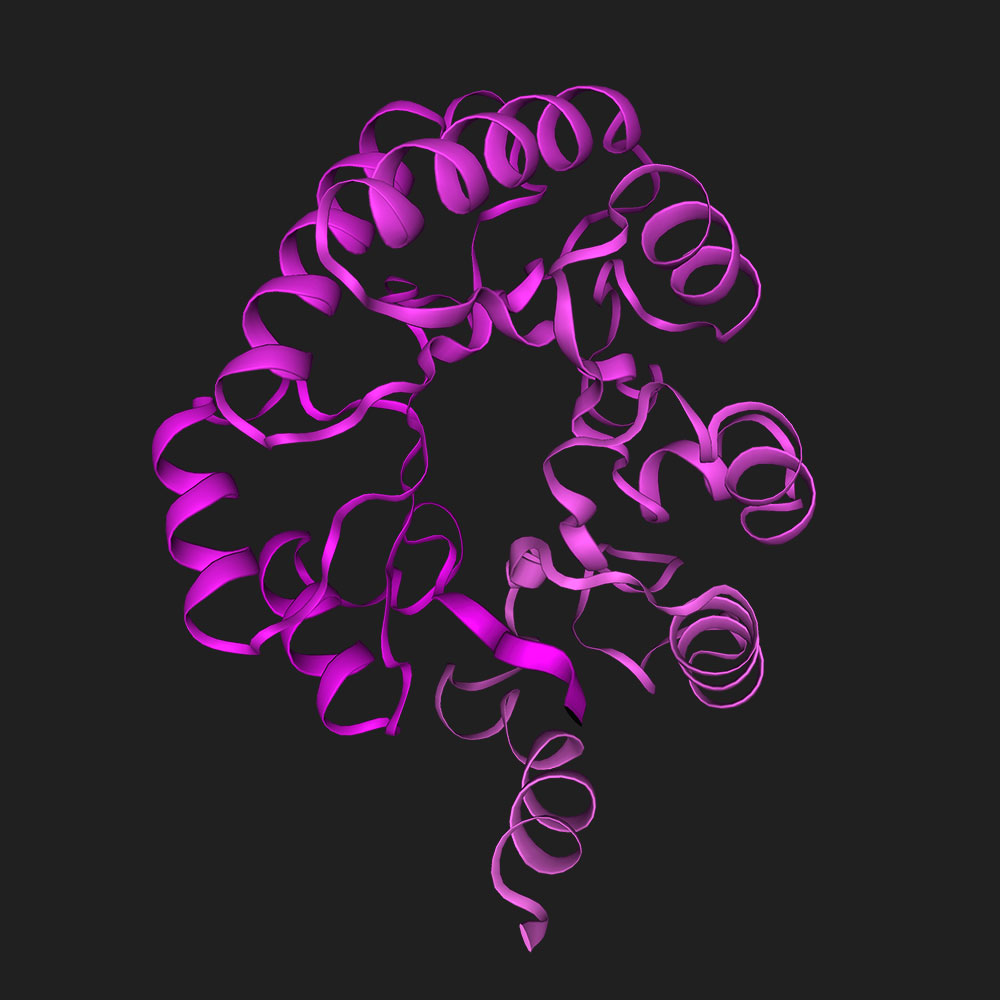
Fold 'GIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKP'
Inputs
Protein sequence: A string of the single letter amino acid sequence. Common 20 amino acids only. For single chain (monomeric) proteins.
Outputs
PDB file: File containing the predicted 3D coordinates of each residue in the input sequence.
pLDDT: Specified in the b-factor column of the PDB file. A per residue confidence score between 0 and 100 with higher being better.
Evaluate Protein Structure Predictions
- pLDDT: Predicted local distance difference test (pLDDT) is a per residue confidence score. 0-50: very low (correlated with disorder/flexibility), 50-70: low, 70-90: high, 90-100: very high (highly structured/stable).
- PAE: Predicted aligned error (PAE) is a residue pair confidence. 0-5 angstroms (low): The relative position between the 2 residues is known (they move together). 20+ anstroms (high): The relative position is not known (they move independently of each other)