AF2BIND for Prediction of Ligand-Binding Sites
AF2BIND is a logistic regression model on top of AF2 embeddings that predicts the probability that each residue in a protein is in a pocket. Binding site prediction is the problem of detecting where (which residues) on a protein are likely to interact with other molecules. This is also called a binding pocket, pocket, functional site, or sometimes a hotspot. Not all parts of a protein can be easily bound, so identifying where likely sites are can help scientists understand what the function of an unknown protein is, avoid disrupting important sites when engineering a protein, and decide whether or not a candidate protein is a good drug target.
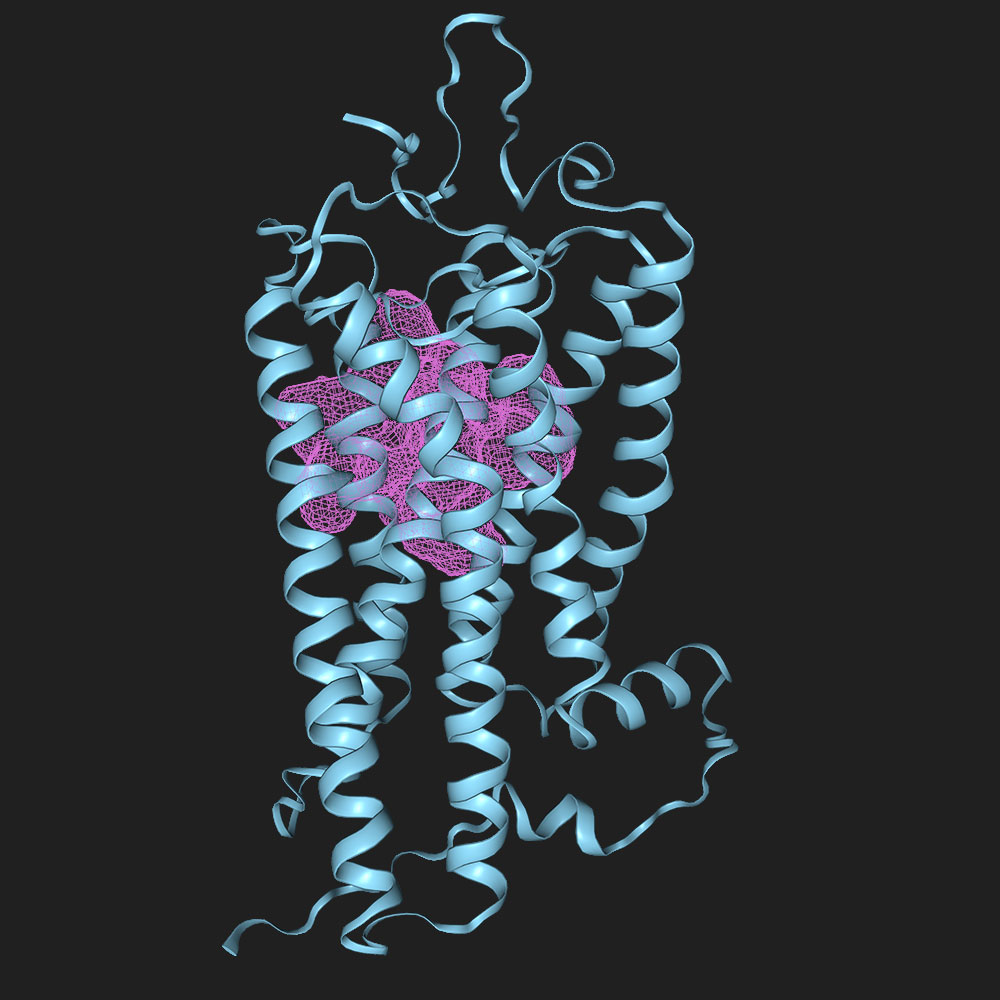
Predict pockets in P08100
| resi | p(bind) |
|---|---|
| 265 | 0.925 |
| 268 | 0.8773 |
| 212 | 0.8214 |
Inputs
PDB file: A single chain protein structure.
Outputs
Probabilities: For each residue, a probability of being in a pocket.
Examples
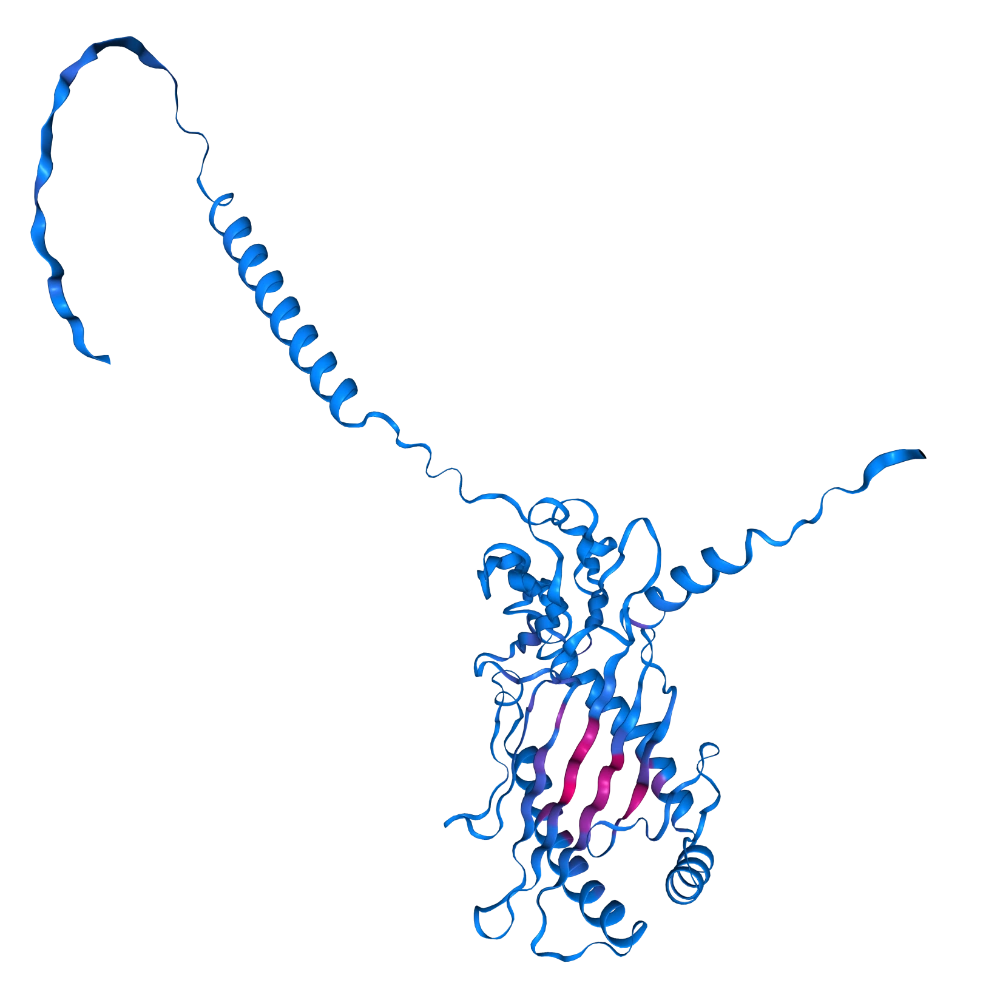
Predict binding pockets of P01889
| resi | p(bind) |
|---|---|
| 33 | 0.8715 |
| 123 | 0.8211 |
| 31 | 0.7818 |
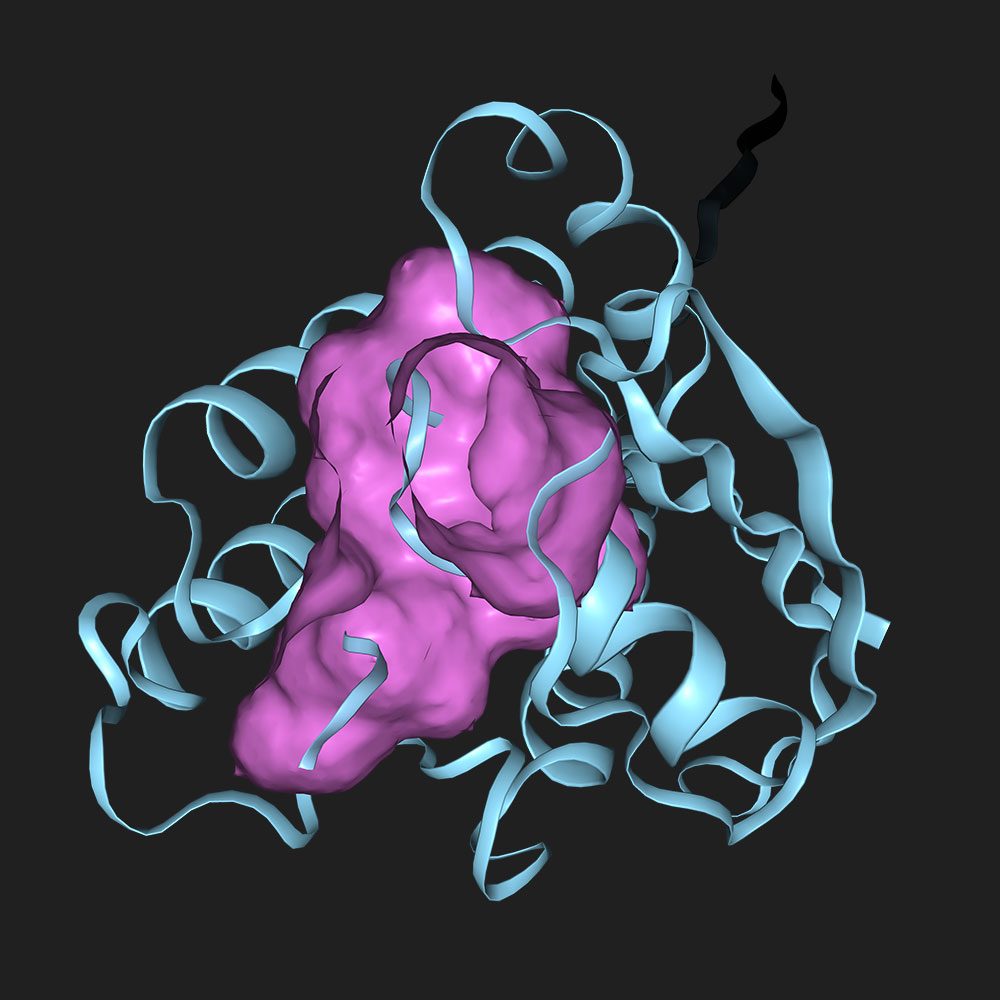
Predict binding sites in P32883
| resi | p(bind) |
|---|---|
| 13 | 0.8182 |
| 15 | 0.8126 |
| 16 | 0.811 |
Analyzing Binding Site Prediction Results
- Visual Inspection: Use of a molecule visualizer with expert knowledge to check if the predicted pocket has sufficient shape and biochemical properties to be a binding surface.
- Docking: If a hypothetical small molecule is known, docking can be used to cross-validate pocket predictions.