DiffDock for Protein-Small-Molecule Docking
DiffDock is a diffusion-based ML model for the docking of a protein to a small molecule. DiffDock reports a confidence score (-inf, inf) where a larger number is better, and scores below -1.5 can be considered low confidence. Note the confidence score does not predict the affinity, aka strength of the biochemical attraction, of the complex.
DiffDock Examples and Docking Scripts
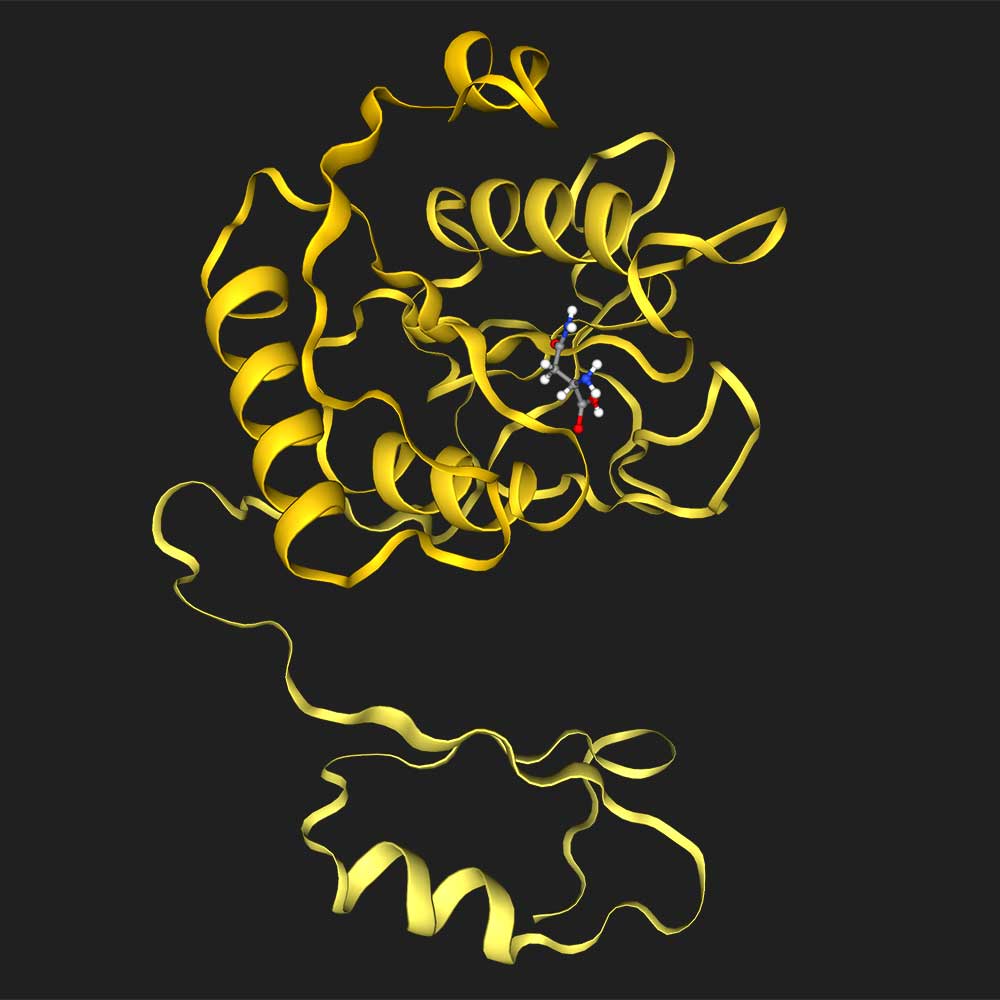
Dock asparagine into A0A1L9RXX7
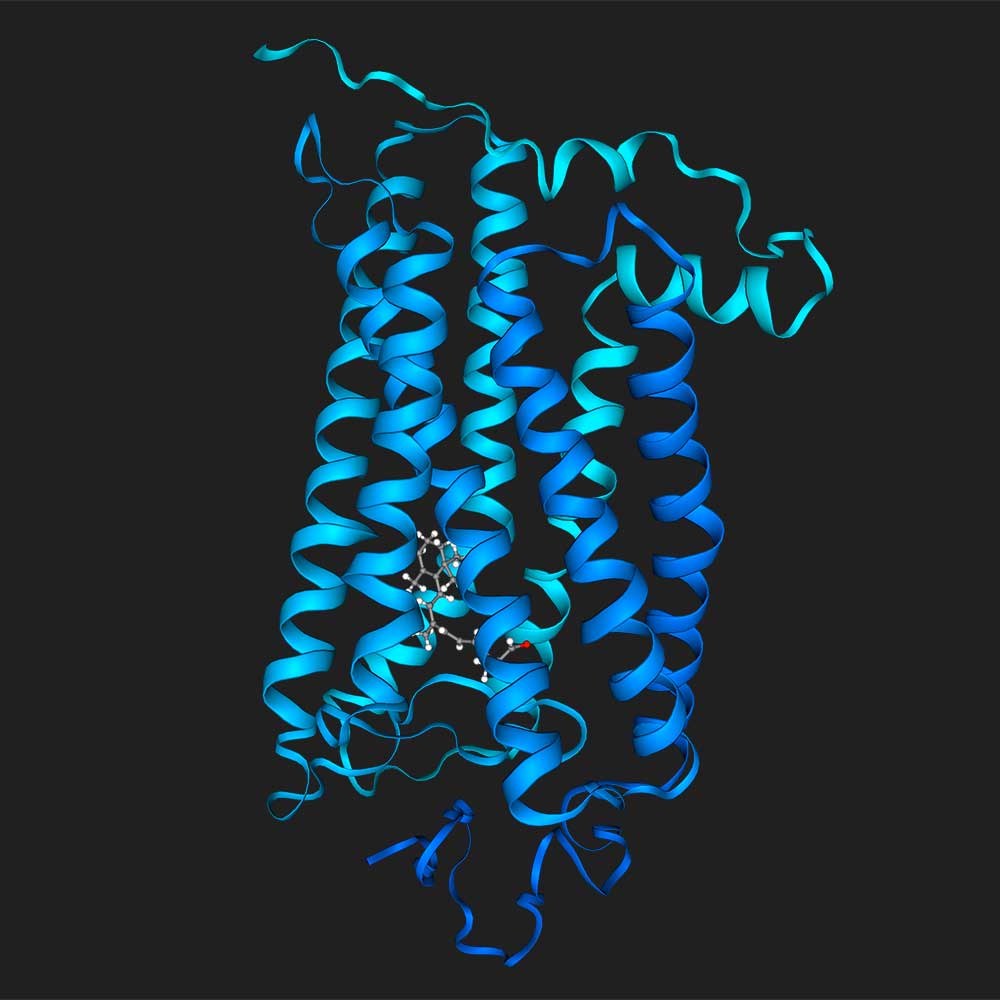
Load the protein structure for uniprot ID P08100, load the small molecule retinal, and then dock the two together
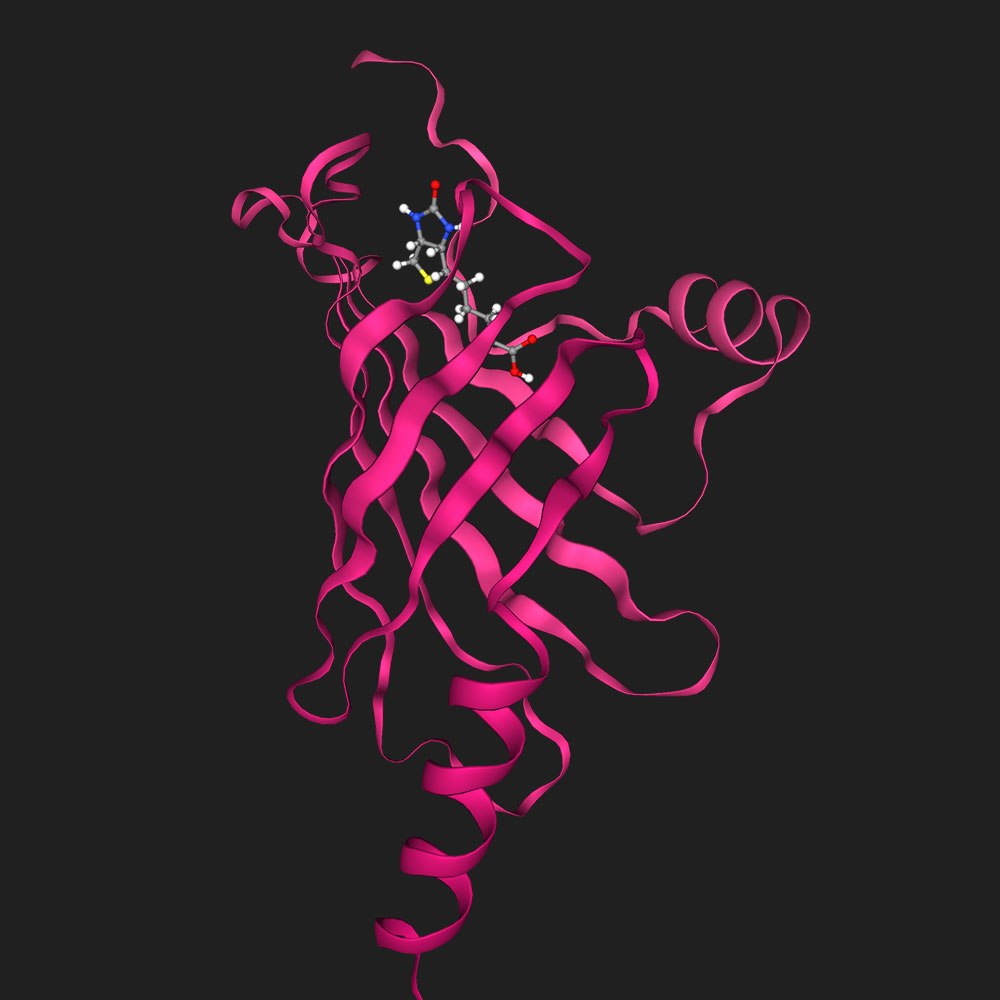
Dock biotin into P22629
Inputs
PDB file: specifies the protein structure in 3D.
SMILE string: specifies the identity of the small molecule.
Outputs
PDB file: specifies the predicted structure of the protein + small molecule docked together.
Confidence score: model confidence in the dock
Evaluating Protein-Small-Molecule Docking Results
High quality computational evaluation of docking results is an underdeveloped area.
- Comparison to Experiment: Alignment to and root-mean-square deviation (RMSD) evaluation of the docked structure to experimental data would be the gold-standard metric. However, in the vast majority of cases experimental data is unavailable.
- Visual Inspection: Use of a molecule visualizer with expert knowledge to check if docked structure makes sense with biochemistry or data that is not a full experimental structure. For example, checking if there is a strong bonding pattern and shape complementarity at the interface. Or for example, checking if the interface involves residues known to be important for the interaction, perhaps from mutational scanning experiments.